Assignment: Running a Random Forest
19 Jan 2017Program and outputs
Data loading and cleaning
import pandas as pandas
from sklearn.model_selection import train_test_split
from sklearn.ensemble import ExtraTreesClassifier, RandomForestClassifier
import sklearn.metrics
import numpy as np
import matplotlib.pylab as plt
CSV_PATH = 'gapminder.csv'
data = pandas.read_csv(CSV_PATH)
print('Total number of countries: {0}'.format(len(data)))
Total number of countries: 213
PREDICTORS = [
'incomeperperson', 'alcconsumption', 'armedforcesrate',
'breastcancerper100th', 'co2emissions', 'femaleemployrate',
'hivrate', 'internetuserate', 'oilperperson',
'polityscore', 'relectricperperson', 'suicideper100th',
'employrate', 'urbanrate'
]
clean = data.copy()
clean = clean.replace(r'\s+', np.NaN, regex=True)
clean = clean[PREDICTORS + ['lifeexpectancy']].dropna()
for key in PREDICTORS + ['lifeexpectancy']:
clean[key] = pandas.to_numeric(clean[key], errors='coerce', downcast='integer')
clean = clean.astype(int)
predictors = clean[PREDICTORS]
targets = clean.lifeexpectancy
pred_train, pred_test, tar_train, tar_test = train_test_split(predictors, targets, test_size=.4)
print(pred_train.shape, pred_test.shape, tar_train.shape, tar_test.shape)
(33, 14) (23, 14) (33,) (23,)
classifier=RandomForestClassifier(n_estimators=25)
classifier=classifier.fit(pred_train,tar_train)
predictions=classifier.predict(pred_test)
print('Confusion matrix:', sklearn.metrics.confusion_matrix(tar_test, predictions), sep='\n')
print('Accuracy score:', sklearn.metrics.accuracy_score(tar_test, predictions))
Confusion matrix:
[[0 0 0 0 0 0 0 1 0 0 0 0 0]
[0 0 1 0 0 0 0 0 0 0 0 0 0]
[0 0 0 0 1 0 0 0 0 0 0 0 0]
[0 0 0 0 1 0 1 0 0 0 0 0 0]
[0 0 1 0 3 0 2 0 0 0 0 0 0]
[0 0 0 0 1 0 0 0 0 0 0 0 0]
[0 0 0 0 0 0 0 0 0 0 0 0 0]
[0 0 0 0 0 0 0 0 0 0 0 0 0]
[0 0 0 0 0 0 0 0 0 0 0 2 0]
[0 0 0 0 0 0 1 0 0 0 0 0 0]
[0 0 0 0 0 0 0 0 0 1 0 4 0]
[0 0 0 0 0 0 0 0 0 0 1 1 0]
[0 0 0 0 0 0 0 0 0 0 0 1 0]]
Accuracy score: 0.173913043478
from table import table
# fit an Extra Trees model to the data
model = ExtraTreesClassifier()
model.fit(pred_train, tar_train)
# display the relative importance of each attribute
importances = list(zip(PREDICTORS, model.feature_importances_))
importances = sorted(importances, key=lambda x: x[1], reverse=True)
table(importances)
| urbanrate | 0.126317199412 |
| breastcancerper100th | 0.105117665487 |
| incomeperperson | 0.0948448657848 |
| internetuserate | 0.0771414112204 |
| employrate | 0.0757854973973 |
| suicideper100th | 0.073923975227 |
| alcconsumption | 0.0678999029417 |
| armedforcesrate | 0.0631145170619 |
| polityscore | 0.0601258137948 |
| co2emissions | 0.0597650190565 |
| femaleemployrate | 0.0555015579106 |
| oilperperson | 0.0544900713322 |
| relectricperperson | 0.0449603576248 |
| hivrate | 0.041012145749 |
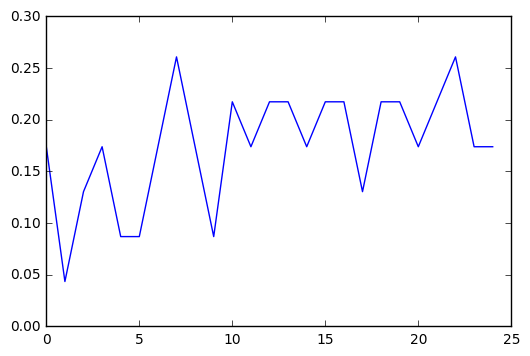
trees = range(25)
accuracy = np.zeros(25)
for idx in trees:
classifier = RandomForestClassifier(n_estimators=idx + 1)
classifier = classifier.fit(pred_train,tar_train)
predictions = classifier.predict(pred_test)
accuracy[idx] = sklearn.metrics.accuracy_score(tar_test, predictions)
plt.cla()
plt.plot(trees, accuracy)
Summary
The gapminder dataset continues to be fairly unimpressive with this form of analysis. The accuracy score of 0.17 indicates that the random forest has very little predictive power for determining life expectancy. I think there just aren’t enough remaining datapoints once the gapminder dataset has been cleaned up.
Urban rate, incidences of breast cancer, and income per person were the most important predictors, but having run the analysis a couple times I found that those numbers varied pretty wildly, nothing consistently stood out as a predictor.
Running the classifier with different numbers of trees was interesting. There was a general curve of improvement to accuracy, which seemed to level off around 10, but there’s enough variance even in that that I don’t think any meaningful conclusions can be drawn.